Single-cell Raman-based tool for efficient mining of live functional microbes from nature
Currently, the best way for scientists to isolate a specific microbe with a particular metabolic function from an environment is to take a sample of cells, culture them, and then screen them for the desired cell functions.
This method has several limitations. First, scientists have not identified most cells in nature, limiting the results to previously cultured cells. Second, the way cells behave in a test tube is not necessarily how they behave in nature, or in situ. This can make it difficult to identify the right cell with the right metabolic functions.
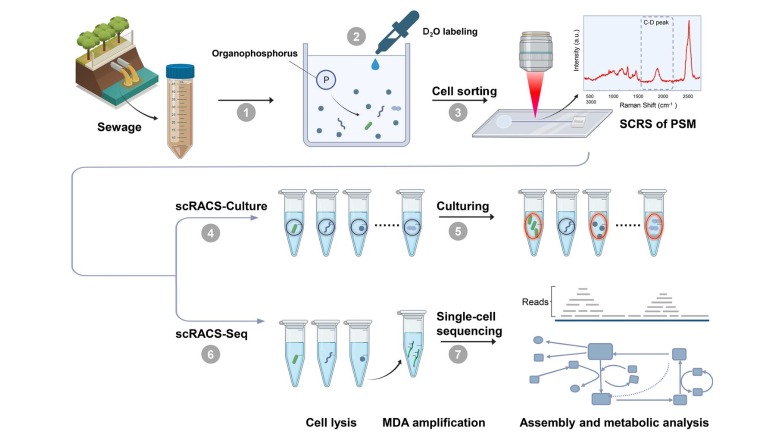
Researchers from the Qingdao Institute of Bioenergy and Bioprocess Technology (QIBEBT) of the Chinese Academy of Sciences (CAS) have proposed a new technique called single-cell Raman-activated sorting and cultivation, or scRACS-Culture, to find and harvest cells from the environment by screening the cells first and then culturing them.
The study was published in ISME Communications on Oct. 30.
"Function-based mining of microbes from nature has traditionally employed a 'culture first, screen second' strategy, which has several limitations," says Jing Xiaoyan, paper first-author and a senior engineer from Single-Cell Center of QIBEBT. "By employing phosphate-solubilizing microbes as a model, we introduced a 'screen first' strategy called single-cell Raman-activated sorting and cultivation."
Using a novel instrument called RACS-Seq, the researchers demonstrated the feasibility of scRACS-Culture by mining microbes for in situ organic-phosphate solubilizing activity from an urban wastewater treatment plant. This shows a real-world use case for this technology because such organic-phosphate solubilizing strains are highly sought-after resources, as they can fight pollution in water bodies and soil, promote nutrient absorption of crops, improve fertilizer efficiency, and reduce the use of chemical fertilizers.
The researchers used the scRACS-Culture method to mine such cells directly from a wastewater sample, by looking at their metabolic rate when only organic-phosphate substrates are available. They pointed out that such culture-independent, "screen-first" strategy is advantageous in that it can screen all cells in a microbiome, instead of just those cells that can be cultured.
Moreover, this strategy can evaluate the in-situ function of the cell, which is usually more relevant than the function of a pure-culture in a test-tube. It is also applicable to a wide variety of valuable metabolic functions of cells. "Because this technique can measure a wide range of metabolic phenotypes in a fluorescence-probe-free manner, it should greatly expand the use of function-driven single-cell technologies in microbiome science and industries," says co-first author Gong Yanhai, assistant research fellow at Single-Cell Center of QIBEBT.
To improve the success rate of scRACS-Culture, they envision a strategy that unlocks the target cell's nutrient needs via metabolic reconstruction of its single-cell genome. The knowledge can then be exploited to optimize the culture medium to grow these yet-to-culture single-cells into valuable live-bacteria cultures, for animal and plant health and for environmental remediation.
Looking ahead, the team is planning to further elevate the scale and throughput of the technology, for efficiently mining the "probiotics" from a wide range of ecosystems, according to Prof. Xu Jian from Single-Cell Center of QIBEBT, who led the study.
More information: Xiaoyan Jing et al, Single-cell Raman-activated sorting and cultivation (scRACS-Culture) for assessing and mining in situ phosphate-solubilizing microbes from nature, ISME Communications (2022). DOI: 10.1038/s43705-022-00188-3.



